SAMP: Send results to other VO tools
SAMP is a VO protocol that allows to share data between VO applications. It was initially designed to work between desktop applications but, thanks to Mark Taylor's sampjs library VOSA, being a web application, can share results with desktop applications too.
Thus, apart from just visualizing and downloading the final results, most of the tables can be broadcasted to any other VO application that is open in the final user computer and connected to the SAMP Hub. In particular, this is specially useful to send some results tables to Topcat for further analysis.
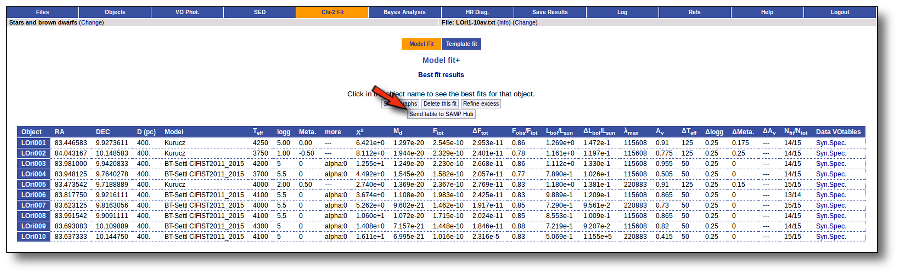
As far as you have an active SAMP Hub in your computer (for instance, if you have Topcat open) you will see a "Sent table to SAMP Hub" button in some of the VOSA results. For example, in a model fit, you can see it:
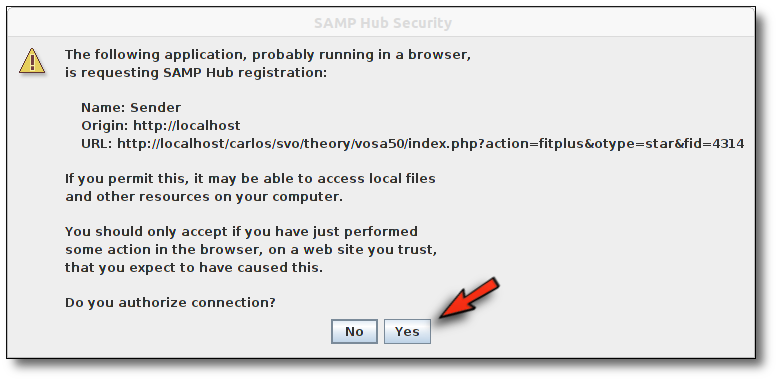
The SAMP Hub will request authorization to broadcast the file sent by VOSA: You can accept it safely.
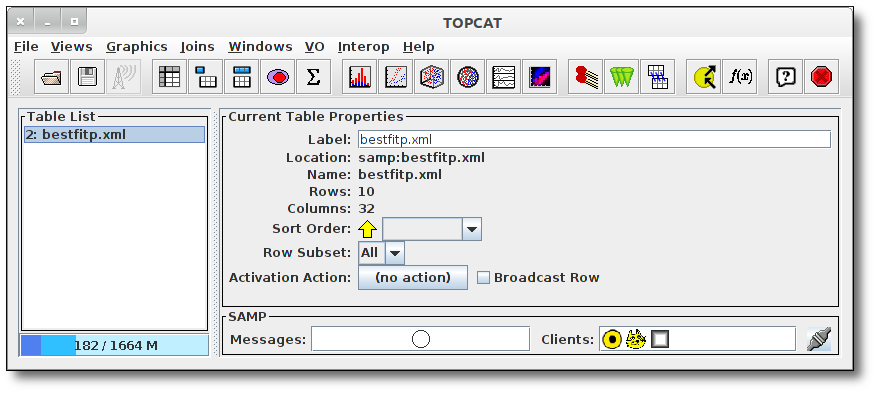
The VO table sent by VOSA will be loaded into TopCat,
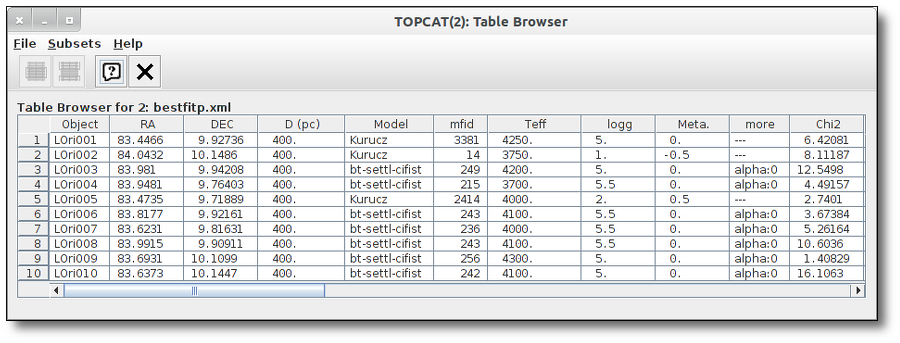
and you will be able to use Topcat functionalities to work with it.
| 
